Advanced Photon Source¶
At synchrotron facilities using the EPICS [B1] software for area detectors [B12] with the NDFileHDF5 plugin [B11], is possible to save Data Exchange files by properly configure the detector and the HDF schema attribute files to obtain txm.h5
Here are the templates in use at the Advanced Photon Source:
- 2-BM-A/B
Micro Tomography Instrument:
hdf_schema.xmlplusA_station_detector_attributes.xmlorB_station_detector_attributes.xml
- 6-BM
Micro Tomography Instrument:
TomoScanLayout.xmlandTomoScanDetectorAttributes.xml
- 7-BM
Fast Micro Tomography Instrument:
mct_hdf_schema.xmlandmct_detector_attribute.xml
- 13-BM
Micro-tomography system at 13-BM-D using PG cameras:
tomoLayout.xmlandtomoDetectorAttributes.xml
- 32-ID
Transmission X-Ray Microscope:
hdf_schema.xmlandtxm_detector_attribute.xml.Micro Tomography Instrument:
mct_hdf_schema.xmlandmct_detector_attribute.xml.
XML¶
To check that the areadetector attributes and layout XML contain a set of matching names run:
$ bash
usertxm@txmtwo$ grep -oP 'name=\"\K[^\"]+' TomoScanDetectorAttributes.xml | while read -r line ; do echo -n "$line " ; grep -q "$line" TomoScanLayout.xml && echo true || echo false ; done | grep false
usertxm@txmtwo$ grep -oP 'ndattribute=\"\K[^\"]+' TomoScanLayout.xml | while read -r line; do echo -n "$line "; grep -q "$line" TomoScanDetectorAttributes.xml && echo true || echo false ; done |grep false
To visualize the meta data and the layout of the hdf file use meta cli
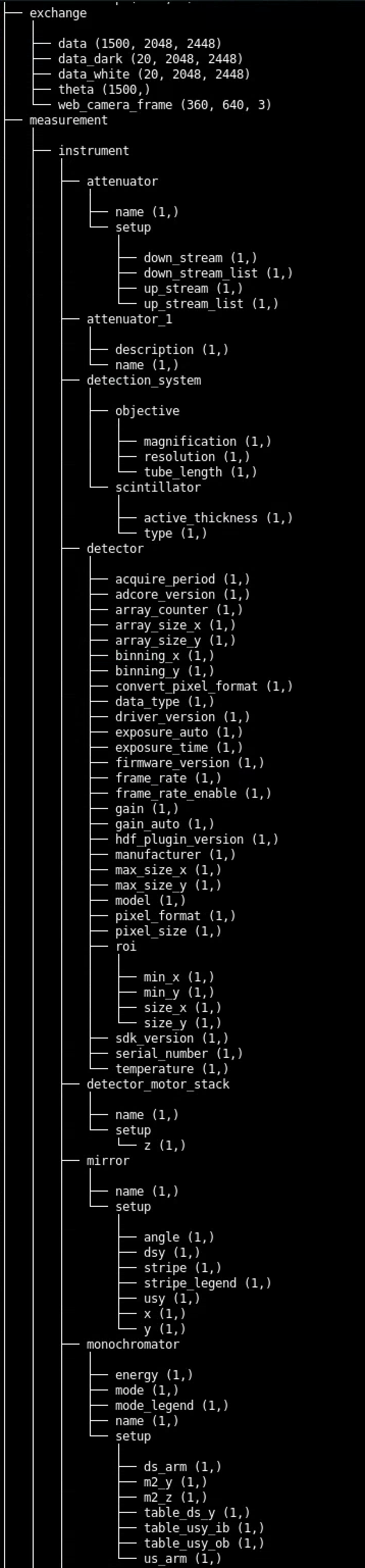
View the hdf tree¶
To view the data tree contained in a generic hdf file:
$ meta tree --file-name data/base_file_name_001.h5
View the meta data¶
To view the meta data contained in a generic hdf file:
$ meta show --file-name data/base_file_name_001.h5
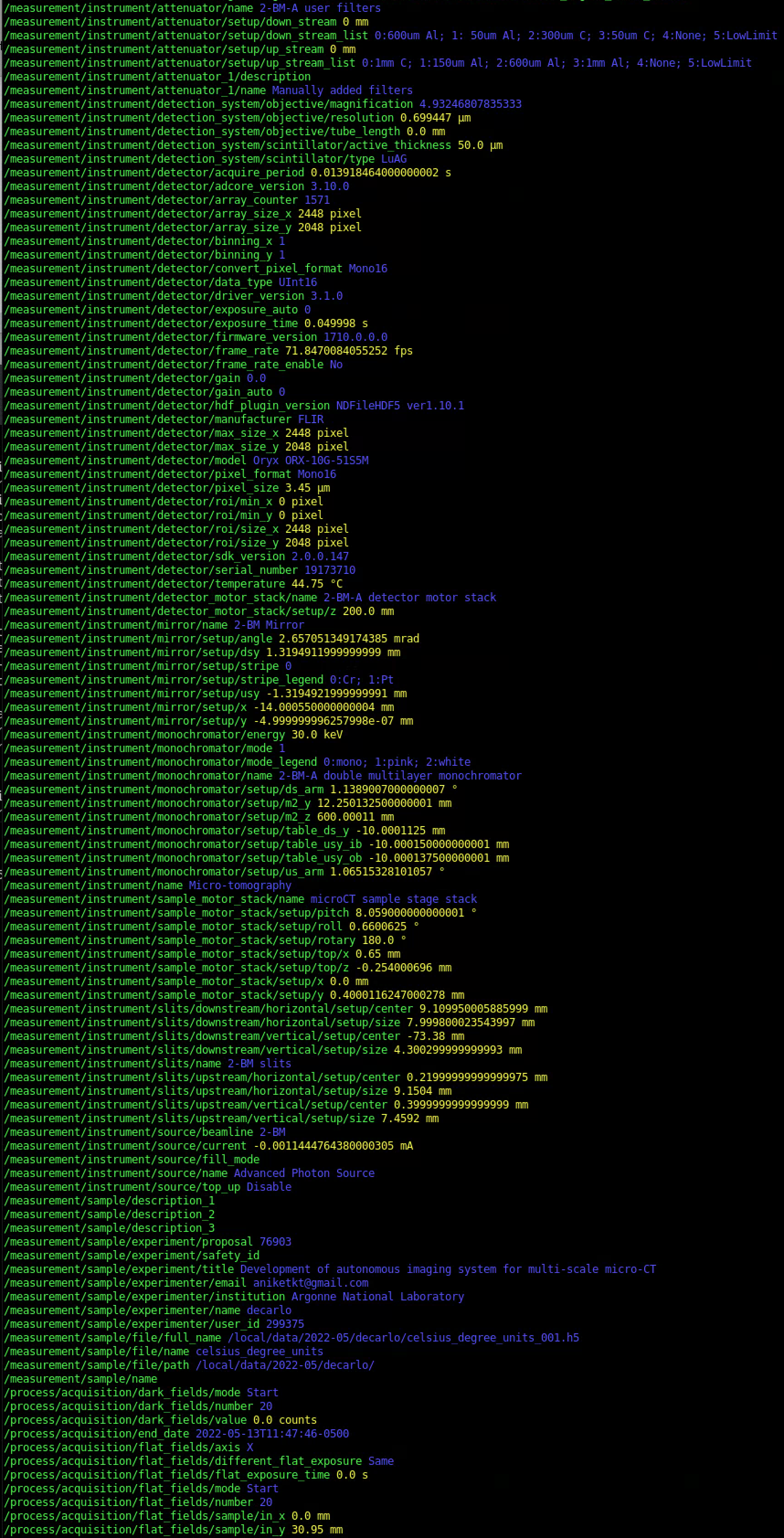
View a subset meta data¶
To view a subset of the meta data contained in a generic hdf file:
$ meta show --file-name data/base_file_name_001.h5 --key energy
Replace an hdf entry value¶
To replace the value of an entry:
$ meta set --file-name data/base_file_name_001.h5 --key /process/acquisition/rotation/rotation_start --value 10
Meta data rst table¶
To generate a meta data rst table compatible with sphinx/readthedocs:
$ meta docs --file-name data/base_file_name_001.h5
2022-02-09 12:30:16,983 - Please copy/paste the content of ./log_2020-05.rst in your rst docs file
The content of the generated rst file will publish in a sphinx/readthedocs document as:
2022-05
decarlo
value |
unit |
|
|---|---|---|
000/measurement/instrument/monochromator/energy |
30.0 |
keV |
000/measurement/instrument/sample_motor_stack/setup/x |
0.0 |
mm |
000/measurement/instrument/sample_motor_stack/setup/y |
0.4000116247000278 |
mm |
000/measurement/sample/experimenter/email |
Note
when using the docs option –file-name can be also a folder, e.g. –file-name data/ in this case all hdf files in the folder will be processed.
to list of all available options:
$ meta -h